
Very few AhpE proteins have been identified and all are found in aerobic gram-positive bacteria of the order Actinomycetales; however, they do not clearly group with any of the other Prx subfamilies (Nelson et al. 2011). The AhpE subfamily includes both 1-Cys and 2-Cys members. Mycobacterium tuberculosis AhpE reacts about two orders of magnitude more slowly with peroxide than members of the AhpC/Prx1 subfamily (~105 vs ~107 M-1s-1, respectively) and the identity of its physiologic reductant is unclear (Hugo et al. 2009).

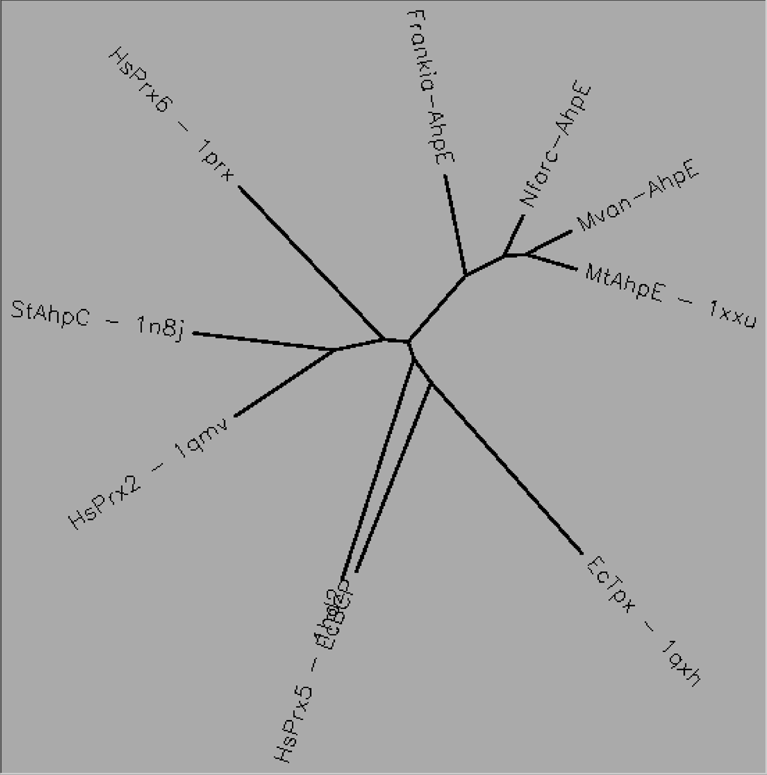
Aligned active site signatures (from DASP; Nelson et al. 2011) from the AhpE subfamily (center group) and one representative each of the other five subfamilies (upper and lower groups). Key residues of the active site signatures are noted by asterisks along the top. Shown below is the unrooted tree generated by Phylip’s Drawtree using the SDSC Biology Workbench tools.
2 structures, 1 protein
| PDB identifiers | Name | Species |
|---|---|---|
| 1xvw, 1xxu | AhpE | Mycobacterium tuberculosis |