
After AhpE, Tpx is the smallest and least phylogenetically diverse Prx subfamily. Tpx subfamily members are all bacterial and are almost exclusively 2-Cys Prxs with the resolving cysteine located on the same subunit of the protein. The prototypical location of the CR (>99% of subfamily members) is in the C-terminal turn of helix α3. For more information about structural transitions between the reduced and oxidized state see (Hall et al. 2009). Although functionally monomeric, all Tpx proteins studied to date form dimers across the A-type interface, apparently independent of redox state. Tpx proteins have been shown to be reduced by thioredoxin. E. coli Tpx (in contrast to E. coli AhpC) is more specific for cumene hydroperoxide (a bulky, hydrophobic substrate) than for hydrogen peroxide (Km of 9 μM compared with 1.7 mM, respectively) (Baker and Poole 2003). Other Tpx proteins have also been shown to react with alkyl hydroperoxides, suggesting that Tpx is important for lipid hydroperoxide removal in bacteria (Cha et al. 2004; Jaeger et al. 2004).
The name Tpx arose from one of the original designations of a “thiol peroxidase”, although this name has also been used more generally to describe Prx proteins found in other subfamilies (Cha et al. 1995). Tpx subfamily members have also been called p20 and have been classified at various times into “group E” (Hofmann et al. 2002), and as part of the “Prx2” grouping (Copley et al. 2004).
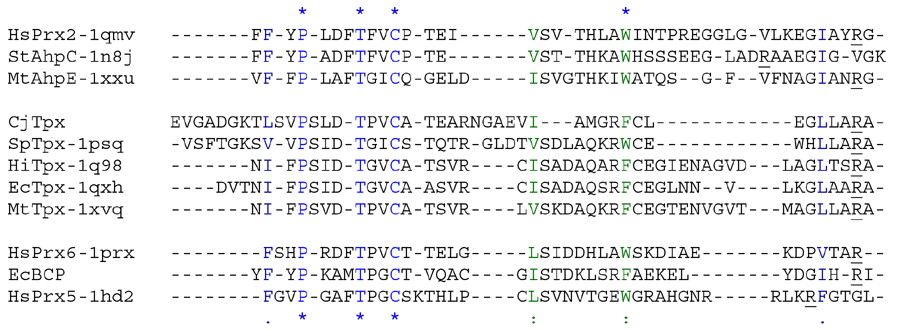
Aligned active site signatures (from DASP; Nelson et al. 2011) from the Tpx subfamily (upper group) and one representative each of the other five subfamilies (lower group). Key residues of the active site signatures are noted by asterisks along the top. Shown below is the unrooted tree generated by Phylip’s Drawtree using the SDSC Biology Workbench tools.
12 structures, 6 proteins
| PDB identifiers | Name | Species |
|---|---|---|
| 1psq | Thiol peroxidase | Streptococcus pneumoniae |
| 1q98 | Thiol peroxidase | Haemophilus influenzae |
| 1qxh, 3hvv, 3hvs, 3i43, 3hvx | Thiol peroxidase | Escherichia coli |
| 1xvq, 1y25 | Tpx | Mycobacterium tuberculosis |
| 2yzh | Thiol peroxidase | Aquifex aeolicus |
| 2jsz, 2jsy | Tpx | Bacillus subtilis |
| 3keb | Thiol peroxidase | Chromobacterium violaceum |