
The AhpC-Prx1 subfamily is essentially synonymous with the “ typical 2-Cys Prxs ” and has also been referred to as the “A” group in the plant field (Trivelli et al. 2003). This subfamily is both the largest and the most widely distributed, with members found in archaea, bacteria, and all classes of eukaryotes (Nelson et al. 2011). Members of this subfamily include the bacterial AhpC proteins, tryparedoxin peroxidases, plant Prxs including Arabidopsis thaliana 2-Cys Prxs and barley Bas1, the yeast TSA1 and TSA2 proteins, and human PrxI, II, III, and IV. In addition to the common core Prx structure, members of this subfamily have a C-terminal extension that contains the CR, which forms a disulfide bond with the CP in its partner subunit across the B-type interface (Hall et al. 2010). For almost all proteins, the CR is conserved in the same position as C172 in human PrxII. An exception is Mycobacterium tuberculosis Prx1, which contains two Cys residues; although Cys174 is considered the primary CR (equivalent to C172 in human PrxII), C176 is also able to function as CR in the C174S mutant (Koshkin et al. 2003). AhpC/Prx1 and Prx6 subfamily members have also been shown to further associate to form octamers, decamers, and even dodecamers across the A-type interface interface in a redox-sensitive manner (Hall et al. 2010; Parsonage et al. 2005).
Although members of this subfamily react very efficiently with hydrogen peroxide (~107 M-1s-1), some (mostly eukaryotic) members of this subfamily are overoxidized at high peroxide concentration to form sulfinic (-SPO2H) and sulfonic (-SPO3H) acids. The sulfinic acid form of some Prxs can be reduced and reactivated by sulfiredoxin (Srx) in an ATP-dependent manner (Jönsson and Lowther 2007). The overoxidized species of some Prx1-like proteins have been shown to form larger aggregates that have molecular chaperone activity (Hall et al. 2009; Jang et al. 2004; Phalen et al. 2006). Members of the Prx1 subfamily have been linked with important roles in cellular signaling and some appear to be regulated by phosphorylation (Rhee et al. 2005; Woo et al. 2010). For a review of structures from the Prx1 subfamily, see (Hall et al. 2010).
In the 2008 version of the database the AhpC-Prx1 subfamily was divided into four groups.
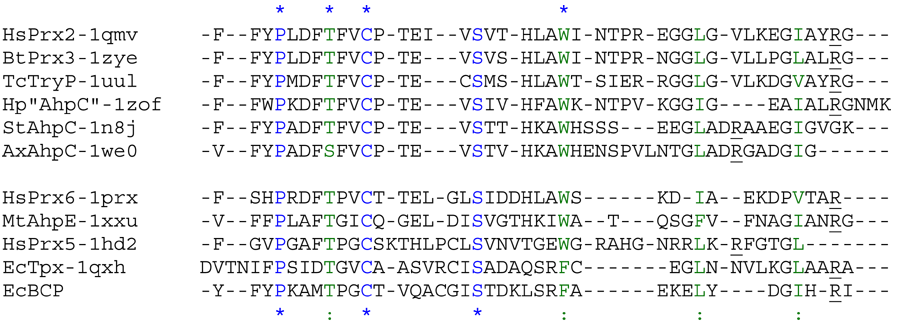
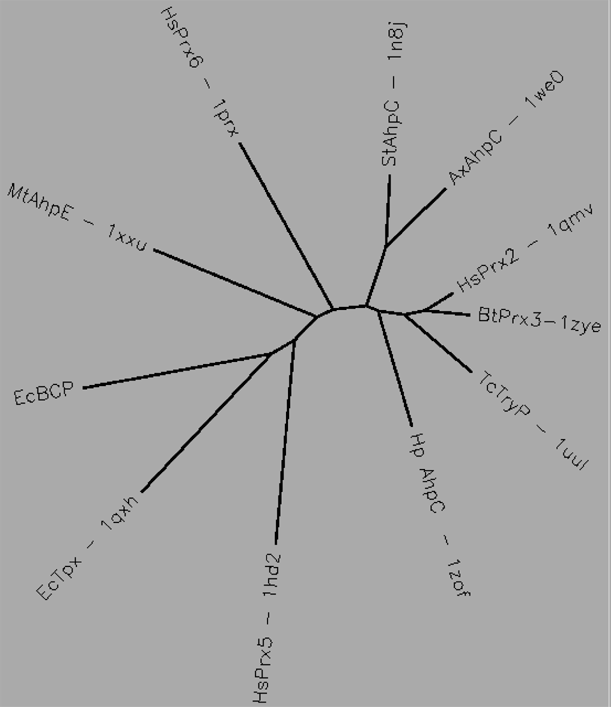
Aligned active site signatures (from DASP; Nelson et al. 2011) from the AhpC/Prx1 subfamily (upper group) and one representative each of the other five subfamilies (lower group). Key residues of the active site signatures are noted by asterisks along the top. Shown below is the unrooted tree generated by Phylip’s Drawtree using the SDSC Biology Workbench tools.
22 structures, 14 proteins
| PDB identifiers | Name | Species |
|---|---|---|
| 1e2y | tryparedoxin peroxidase | Crithidia fasciculata |
| 1qmv | Prx2 | Homo sapiens |
| 1qq2, 2z9s | Prx1 | Rattus norvegicus |
| 1uul | tryparedoxin peroxidase | Trypanosoma cruzi |
| 1zof | AhpC | Helicobacter pylori |
| 1zye | Prx3 | Bos taurus |
| 2h01 | thiol peroxidase 1 | Plasmodium yoelli |
| 2c0d | Mitochondrial 2-Cys Prx | Plasmodium falciparum |
| 2pn8 | Prx4 | Homo sapiens |
| 2rii, 2hy2 | Prx1 (complex with Srx) | Homo sapiens |
| 2h66, 2i81 | 2-Cys | Plasmodium vivax |
| 1yep, 1n8j, 1yex, 1yfo, 1yf1, 3emp | AhpC | Salmonella typhimurium |
| 1we0 | AhpC | Amphibacillus xylanus |
| 2bmx | AhpC | Mycobacterium tuberculosis |