Research Interests
Mammalian RNase H2
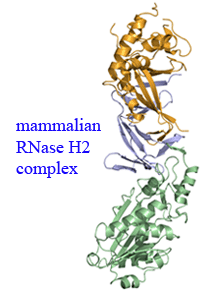 The mammalian RNase H2 ribonuclease complex has a critical function in nucleic acid metabolism to prevent immune activation with likely roles in processing of RNA primers in
Okazaki fragments during DNA replication, in removing ribonucleotides misinserted by DNA polymerases, and in eliminating RNA:DNA hybrids during cell death. Mammalian RNase H2
is a heterotrimeric complex of the RNase H2A, RNase H2B, and RNase H2C proteins that are all required for proper function and activity. Mutations in the human RNase H2 genes
cause Aicardi-Goutieres syndrome (AGS). We have determined the crystal structure of the three-protein mouse RNase H2 enzyme complex to better understand the molecular basis of
RNase H2 dysfunction in human autoimmunity. The structure reveals the intimately interwoven architecture of RNase H2B and RNase H2C that interface with RNase H2A in a complex
ideally suited for nucleic acid binding and hydrolysis coupled to protein-protein interaction motifs that could allow for efficient participation in multiple cellular functions.
The mammalian RNase H2 ribonuclease complex has a critical function in nucleic acid metabolism to prevent immune activation with likely roles in processing of RNA primers in
Okazaki fragments during DNA replication, in removing ribonucleotides misinserted by DNA polymerases, and in eliminating RNA:DNA hybrids during cell death. Mammalian RNase H2
is a heterotrimeric complex of the RNase H2A, RNase H2B, and RNase H2C proteins that are all required for proper function and activity. Mutations in the human RNase H2 genes
cause Aicardi-Goutieres syndrome (AGS). We have determined the crystal structure of the three-protein mouse RNase H2 enzyme complex to better understand the molecular basis of
RNase H2 dysfunction in human autoimmunity. The structure reveals the intimately interwoven architecture of RNase H2B and RNase H2C that interface with RNase H2A in a complex
ideally suited for nucleic acid binding and hydrolysis coupled to protein-protein interaction motifs that could allow for efficient participation in multiple cellular functions.
TREX1 3'-exonuclease
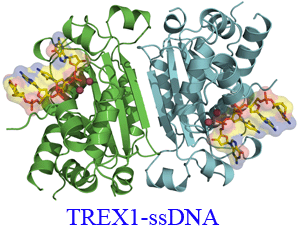 The TREX1 enzyme processes DNA ends as the major 3'- 5' exonuclease activity in human cells. Mutations in the TREX1 gene are an underlying cause of the neurological brain disease
Aicardi-Goutieres syndrome implicating TREX1 dysfunction in an aberrant immune response. TREX1 action during apoptosis likely prevents autoimmune reaction to DNA that would otherwise persist.
To understand the impact of TREX1 mutations identified in patients with Aicardi-Goutieres syndrome on structure and activity we determined the X-ray crystal structure of the dimeric mouse
TREX1 protein in substrate and product complexes containing single-stranded DNA and deoxyadenosine monophosphate, respectively. The structures show the specific interactions between the
bound nucleotides and the residues lining the binding pocket of the 3' terminal nucleotide within the enzyme active site that account for specificity, and provide the molecular basis
for understanding mutations that lead to disease.
The TREX1 enzyme processes DNA ends as the major 3'- 5' exonuclease activity in human cells. Mutations in the TREX1 gene are an underlying cause of the neurological brain disease
Aicardi-Goutieres syndrome implicating TREX1 dysfunction in an aberrant immune response. TREX1 action during apoptosis likely prevents autoimmune reaction to DNA that would otherwise persist.
To understand the impact of TREX1 mutations identified in patients with Aicardi-Goutieres syndrome on structure and activity we determined the X-ray crystal structure of the dimeric mouse
TREX1 protein in substrate and product complexes containing single-stranded DNA and deoxyadenosine monophosphate, respectively. The structures show the specific interactions between the
bound nucleotides and the residues lining the binding pocket of the 3' terminal nucleotide within the enzyme active site that account for specificity, and provide the molecular basis
for understanding mutations that lead to disease.
AlkB DNA demethylase
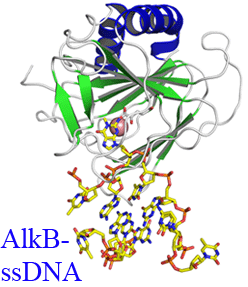 In E. coli cytotoxic DNA methyl lesions on the N1 position of purines and N3 position of pyrimidines are primarily repaired by the 2-oxoglutarate (2-OG) iron(II) dependent dioxygenase, AlkB.
AlkB repairs 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) lesions, but it also repairs 1-methylguanine (1-meG) and 3-methylthymine (3-meT) at a much less efficient rate. How the AlkB
enzyme is able to locate and identify methylated bases in ssDNA has remained an open question. We have determined the crystal structures of the Escherichia coli AlkB protein holoenzyme and the AlkB-ssDNA complex containing a 1-methylguanine lesion. We have coupled this to site
directed mutagenesis of amino acids in and around the active site and tested the effects of these mutations on the ability of the protein to bind both damaged and undamaged DNA as well as catalyze repair of a methylated substrate.
A comparison of our substrate bound AlkB-ssDNA complex with our unliganded holoenzyme reveals conformational changes of residues within the active site that are important for binding damaged bases. Site directed
mutagenesis of these residues reveals novel insight into their roles in DNA damage recognition and repair. Our data support a model that the AlkB protein utilizes at least two distinct conformations in searching and binding methylated bases within DNA, a 'searching' and 'repair' mode. Moreover, we are able to functionally separate these modes through mutagenesis of residues that affect one or the other binding state.
In E. coli cytotoxic DNA methyl lesions on the N1 position of purines and N3 position of pyrimidines are primarily repaired by the 2-oxoglutarate (2-OG) iron(II) dependent dioxygenase, AlkB.
AlkB repairs 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) lesions, but it also repairs 1-methylguanine (1-meG) and 3-methylthymine (3-meT) at a much less efficient rate. How the AlkB
enzyme is able to locate and identify methylated bases in ssDNA has remained an open question. We have determined the crystal structures of the Escherichia coli AlkB protein holoenzyme and the AlkB-ssDNA complex containing a 1-methylguanine lesion. We have coupled this to site
directed mutagenesis of amino acids in and around the active site and tested the effects of these mutations on the ability of the protein to bind both damaged and undamaged DNA as well as catalyze repair of a methylated substrate.
A comparison of our substrate bound AlkB-ssDNA complex with our unliganded holoenzyme reveals conformational changes of residues within the active site that are important for binding damaged bases. Site directed
mutagenesis of these residues reveals novel insight into their roles in DNA damage recognition and repair. Our data support a model that the AlkB protein utilizes at least two distinct conformations in searching and binding methylated bases within DNA, a 'searching' and 'repair' mode. Moreover, we are able to functionally separate these modes through mutagenesis of residues that affect one or the other binding state.
AmrZ transcription factor
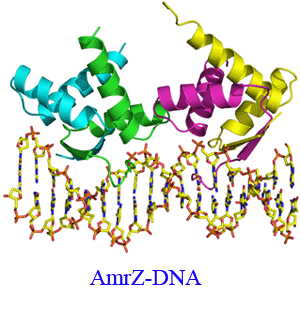
Pseudomonas aeruginosa is a common opportunistic pathogen which is a leading cause of nosocomial infections and chronic lung infections in patients with the autosomal recessive disease cystic fibrosis. In order to establish chronic infections, P. aeruginosa utilizes a multitude of virulence factors which give the bacteria a selective advantage. These virulence factors include the biosynthesis of the exopolysaccharide alginate, repression of flagellum biosynthesis, biogenesis of type IV pili, and repression of its own transcription. The transcription factor AmrZ has dual roles as both an activator and repressor of genes leading to each of these unique phenotypes. The crystal structure of the AmrZ protein in complex with the 18bp repression site on the amrZ gene illuminates the important interactions in the ribbon-helix-helix domain necessary for DNA recognition. We have performed truncation mutagenesis, along with biochemical, biophysical and in vivo experiments to determine that the C-terminal domain of AmrZ is responsible for the formation of oligomeric structures beyond the dimeric state and how these structures regulate gene expression.
Department of Biochemistry and Center for Structural Biology
Wake Forest University School of Medicine
Medical Center Blvd
Winston-Salem, NC 27157
Tel (336) 716.0768
Fax (336) 777.3242
Email: thollis@wfubmc.edu,
